'Variational Graph Autoencoder loss get nan(tensorflow implementation)
I'm implementing VGAE(Variational Graph Autoencoder) in tensorflow.
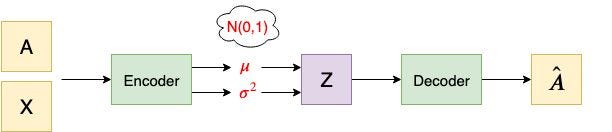
The article has a image of VGAE. I put the VGAE network architecture image here to help you understand what I'm trying to do.
At the first epoch, it shows nan loss value. I assume there is a operation that makes nan value.
However, I can't find the place.
(The code below can be run if you have the imported packages. -> copy and paste)
import tensorflow as tf
from keras import layers, activations
from keras.utils import data_utils
from keras.models import Model
import keras.backend as K
import numpy as np
from keras import metrics, losses, optimizers
import os
import pandas as pd
import networkx as nx
from scipy.sparse import csr_matrix
zip_file = data_utils.get_file(fname="cora.tgz", origin="https://linqs-data.soe.ucsc.edu/public/lbc/cora.tgz", extract=True)
data_dir = os.path.join(os.path.dirname(zip_file), "cora")
citations = pd.read_csv(
os.path.join(data_dir, "cora.cites"),
sep="\t",
header=None,
names=["target", "source"]
)
columns_names = ["paper_id"] + [f"term_{idx}" for idx in range(1433)] + ["subject"]
papers = pd.read_csv(
os.path.join(data_dir, "cora.content"), sep="\t", header=None, names=columns_names
)
class_values = sorted(pd.unique(papers["subject"]))
class_id = {name:id for id, name in enumerate(class_values)}
paper_id = {name: idx for idx, name in enumerate(sorted(pd.unique(papers["paper_id"])))}
citations["source"] = citations["source"].apply(lambda name: paper_id[name])
citations["target"] = citations["target"].apply(lambda name: paper_id[name])
feature_names = set(papers.columns) - {"paper_id", "subject"}
cora_g = nx.from_edgelist(citations.to_numpy())
cora_adj = nx.adjacency_matrix(cora_g)
edges = citations[["source", "target"]].to_numpy().T
edge_weights = tf.ones(shape=edges.shape[1])
node_features = tf.cast(
papers.sort_values("paper_id")[feature_names].to_numpy(), dtype=tf.dtypes.float32
)
graph_info = (node_features, edges, edge_weights, csr_matrix.toarray(cora_adj))
class VGAE(Model):
def __init__(self, encoder, decoder, **kwargs):
super(VGAE, self).__init__(**kwargs)
self.encoder = encoder
self.decoder = decoder
self.total_loss_tracker = metrics.Mean(name="total_loss")
self.reconstruction_loss_tracker = metrics.Mean(name="reconstruction_loss")
self.kl_loss_tracker = metrics.Mean(name="kl_loss")
@property
def metrics(self):
return [
self.total_loss_tracker,
self.reconstruction_loss_tracker,
self.kl_loss_tracker
]
def train_step(self, graph_info):
node_features, adj_matrix = graph_info[0]
with tf.GradientTape() as tape:
z_mean, z_log_var, z = self.encoder([node_features, adj_matrix])
reconstruction = self.decoder(z)
reconstruction_loss = losses.binary_crossentropy(adj_matrix, reconstruction)
kl_loss = losses.KLDivergence()(adj_matrix, reconstruction)
total_loss = reconstruction_loss + kl_loss
grads = tape.gradient(total_loss, self.trainable_weights)
self.optimizer.apply_gradients(zip(grads, self.trainable_weights))
self.total_loss_tracker.update_state(total_loss)
self.reconstruction_loss_tracker.update_state(reconstruction_loss)
self.kl_loss_tracker.update_state(kl_loss)
return {
"loss": self.total_loss_tracker.result(),
"reconstruction_loss": self.reconstruction_loss_tracker.result(),
"kl_loss": self.kl_loss_tracker.result()
}
class GCN(layers.Layer):
def __init__(
self,
output_dim,
activation="relu",
normalize=True,
*args,
**kwargs
):
super(GCN, self).__init__(*args, **kwargs)
self.normalize = normalize
self.output_dim = output_dim
self.activation = activations.get(activation)
def build(self, input_shape):
self.dense = layers.Dense(self.output_dim, kernel_initializer="glorot_uniform")
self.dot = layers.Dot(axes=(1,2))
def call(self, inputs, *args, **kwargs):
node_features, adj_matrix = inputs
if self.normalize:
D = tf.linalg.diag(tf.reduce_sum(adj_matrix, axis=2))
normalized_D = K.pow(D, -0.5)
adj_matrix = self.dot([normalized_D, self.dot([adj_matrix, normalized_D])])
x = self.dense(node_features)
h = self.dot([adj_matrix, tf.transpose(x, perm=[0,2,1])])
return self.activation(h)
def vgae_encoder(input_shapes, output_dim=2):
node_features_shape, adj_matrix_shape = input_shapes
node_feature_input = layers.Input(shape=node_features_shape)
adj_matrix_input = layers.Input(shape=adj_matrix_shape)
h = GCN(output_dim=output_dim, activation="relu", normalize=True, name="gcn_h")([node_feature_input, adj_matrix_input])
z_mean = GCN(output_dim=output_dim, activation="linear", normalize=True, name="gcn_mean")([h, adj_matrix_input])
z_log_var = GCN(output_dim=output_dim, activation="linear", normalize=True, name="gcn_log_var")([h, adj_matrix_input])
_, batch, dim = tf.shape(z_mean)
epsilon = K.random_normal(shape=(batch, dim))
z = z_mean + tf.exp(0.5 * z_log_var) * epsilon
return Model(inputs=[node_feature_input, adj_matrix_input], outputs=[z_mean, z_log_var, z])
def vgae_decoder(num_nodes, output_dim=2):
z = layers.Input(shape=(num_nodes,output_dim))
reconstruction = layers.Dot(axes=(1,2))([tf.transpose(z, perm=[0,2,1]), z])
reconstruction = activations.sigmoid(reconstruction)
return Model(inputs=z, outputs=reconstruction)
encoder = vgae_encoder((node_features.shape, cora_adj.shape), output_dim=2)
decoder = vgae_decoder(len(node_features), output_dim=2)
node_features_expanded = np.expand_dims(node_features, 0).astype(np.float32)
cora_adj_expanded = np.expand_dims(csr_matrix.toarray(cora_adj), 0).astype(np.float32)
vgae = VGAE(encoder, decoder)
vgae.compile(optimizer=optimizers.adam_v2.Adam())
history = vgae.fit((node_features_expanded, cora_adj_expanded), epochs=300, batch_size=1)
Epoch 1/300
1/1 [==============================] - 2s 2s/step - loss: nan - reconstruction_loss: nan - kl_loss: nan
Sources
This article follows the attribution requirements of Stack Overflow and is licensed under CC BY-SA 3.0.
Source: Stack Overflow
| Solution | Source |
|---|
